Compiled by Norris H. Williams, J. Richard Abbott, Kurt Neubig, W. Mark Whitten
Task assignment number SL849#148
5 June 2009 project # 00076531
Abstract
The central premise of DNA barcoding is that each species has a unique set of DNA sequences, of which a carefully chosen subset (a “DNA barcode”) can serve as a baseline reference for comparative identification. Traditionally, plant identification is based on morphology, with the limitation that many species must be collected at a very specific time of the year so that reproductive features can be used for accurate identification. With DNA data, once a reference library is built, barcode identification of plants is theoretically possible at all life stages, from seed to mature plant. Even a fragment of a leaf might be used to identify a species. DNA barcoding has practical utility for all research, wildlife and land management, and conservation efforts that rely on identifying plant species, and it also has applications in forensics, biosecurity, trade in controlled species, foodstuffs, and herbal medicines, and in scientific questions involving evolution, biogeography, and population structure. DNA barcoding is not meant to replace detailed population genetic studies, but it will provide a baseline of genetic data for comparative analysis, and, with sampling of multiple populations, barcoding studies can provide a first glimpse at genetic differences that could reflect underlying patterns of cryptic or incipient species differences. DNA barcoding will enhance our understanding of the Florida flora and should become a useful tool for research and wildlife management. As a prelude of a larger “Barcoding the Flora of Florida” project, we collected and sequenced matK, rbcL, and trnH-psbA spacer for all 136 species on the Florida Exotic Pest Plant Council list of invasive species. With 100% success at generating DNA barcode data for all FLEPPC species and a total DNA sequencing success rate of about 99% (on an individual gene region basis), this study provides compelling evidence that barcoding is logistically feasible. Furthermore, these data generally show excellent differentiation among the invasive species of Florida. However, more data for all species in Florida are required for reliable plant identification by way of DNA barcoding, i.e., the DNA reference library needs to be built for the entire flora.
Introduction
DNA barcoding is currently a hot topic, drawing much excitement for its potential applications. The central premise of DNA barcoding is that each plant species has a unique set of DNA, of which a carefully chosen subset (a “DNA barcode”), can serve as a unique baseline reference for identification.
For this project, we requested funds to barcode the complete list of Florida Exotic Pest Plant Council (FLEPPC) invasive species. The objectives of this project have been: 1) to produce well-vouchered, accurately identified specimens and field-preserved tissues (to ensure high quality DNA) of plant species on the FLEPPC list (a total of 67 spp in Category I and 69 spp in Category II); 2) to provide long-term curation of these voucher specimens; 3) to extract DNA from these specimens and store the DNA in archival conditions, creating a lasting, reliable databank of vouchered DNA samples ultimately to be expanded to include all vascular plant species in Florida; 4) to generate sequence data of three DNA regions (barcodes) for all FLEPPC species, providing insight into the utility of DNA barcoding in Florida; and 5) to analyze and publish these data, with a special emphasis on potential utility for conservation and land management issues, for use by all agencies involved in conservation and wildlife management in Florida.
DNA barcoding has numerous promising uses: identification of different life stages that are often unidentifiable with certainty using traditional morphology, e.g., seeds and seedlings; identification of fragments of plant material; forensics; verification of foodstuffs and herbal medicines; biosecurity and trade in controlled species; and inventories or ecological surveys. Apart from the obvious botanical and genetic research, which are important for our understanding and proper management of the flora, barcoding can directly benefit ecological plot work, which often requires identifying tiny sterile plantlets and conservation or wildlife management work by zoologists or entomologists trying to identify plants (often via gut content or fecal analysis) being used by their study organisms. Thus, there are potential implications for DNA barcoding for all research, management, and conservation efforts that rely on identifying plant species.
Vouchers are plant reference specimens: essential, scientific proof of accuracy for the databank of tissue and DNA that will be used to generate DNA barcode reference data, and they have a lasting research value. A DNA barcoding project must be anchored by voucher specimens and their long-term curation in an herbarium. The UF herbarium (FLAS), part of the Florida Museum of Natural History (FLMNH), is the largest in Florida and represents one of the most complete collections of the flora of Florida. FLAS has the facilities to identify, process, curate, and maintain the voucher specimens. In addition to providing expertise, facilities, and personnel to conduct this project, FLAS & FLMNH are uniquely qualified in that we can store the extracted DNA long-term in our new, NSF-funded, liquid nitrogen tissue & DNA long term storage facility (http://www.floridamuseum.ufl.edu/grr/).
Materials and Methods
Taxon sampling
Specimens were collected from wild-collected and cultivated plants (Table 1). Sampling included all of the species on the Florida Exotic Plant Pest Council list of invasive plant species. In addition, 28 samples (of 27 species) were sampled in duplicate to independently corroborate the validity and accuracy of the DNA data.
Choice of DNA barcode regions
At present, the botanists have not agreed upon a standardized set of DNA regions to use as plant barcodes (see http://www.barcoding.si.edu/plant_working_group.html). Choice of regions involves tradeoffs between universality and ease of amplification/ sequencing vs. DNA variation. Based upon comparisons of Fazekas et al. (2008), we selected three plastid regions: rbcL, matK, and the trnH-psbA intergenic spacer.
Extractions, Amplification and Sequencing
All freshly-collected material was preserved in silica gel (Chase & Hills, 1991). Genomic DNA was extracted using a modified cetyl trimethylammonium bromide (CTAB) technique (Doyle & Doyle, 1987), scaled to a 1 mL volume reaction. Approximately 10 mg of dried tissue were ground in 1 mL of CTAB 2X buffer and 7 µL of proteinase-K at manufacturer’s recommended concentration. Some total DNAs were then cleaned with Qiagen QIAquick PCR purification columns to remove any inhibitory secondary compounds. Amplifications were performed using an Eppendorf Mastercycler EP Gradient S thermocycler and Sigma brand reagents in 25 µL volumes with the following reaction components: 0.5-1.0 µL template DNA (~10-100 ng), 16-17.5 µL water, 2.5 µL 10X buffer, 2-4 µL MgCl2, 0.5 µL of 10 µM dNTPs, 0.5 µL each of 10 µM primers, and 0.5 units Sigma Jumpstart Taq polymerase. All regions were amplified with the parameters 94oC, 3 min; 33X (94oC, 45 sec; 55oC, 45 sec; 72oC, 2 min); 72oC, 3 min. For rbcL, primers rbcLa F (ATGTCACCACAAACAGAGACTAAAGC) and rbcLa R (GTAAAATCAAGTCCACCRCG) were used (Kress and Erickson 2007). For trnH-psbA spacer, primers trnH-psbA F (TGATCCACTTGGCTACATCCGCC) and trnH-psbA R (GCTAACCTTGGTATGGAAGT) from Xu et al. (2000). For matK, primers 390 F (CGATCTATTCATTCAATATTTC) and 1326 R (TCTAGCACACGAAAGTCGAAGT) from Cuenoud et al. (2002). Alternative primers for ferns were used for matK: matK x F (ATACCCCATTTTATTCATCC) and Equisetum R (GTACTTTTATGTTTACGAGC) (http://www.kew.org/barcoding/update.html [site not active])
Products were cleaned using ExoSAP™ (USB Corporation, OH, USA) following the manufacturer’s protocols and were directly sequenced using BigDye terminator reagents on an ABI 3130 automated sequencer according to the manufacturer’s protocols (Applied Biosystems, Foster City, California, USA). Electropherograms were edited and assembled using Sequencher 4.6™ (GeneCodes, Ann Arbor, MI, USA). All sequences are being deposited in GenBank and will be released on 1 September 2009 (Table 1).
Data analysis
Sequence data of matK and rbcL were manually aligned using Se-Al v2.0a11 (Rambaut, 1996). No sequence data were excluded from analyses. Indels (insertions/deletions) were not coded as characters. Analyses were performed using PAUP*4.0b10 (Swofford, 2003) with Fitch parsimony (unordered characters with equal weights; Fitch, 1971) using a heuristic search strategy consisted of branch swapping by tree bisection reconnection (TBR), stepwise addition with 5000 random-addition replicates holding 5 trees at each step, and saving multiple trees (MulTrees). Sequence data were verified, where applicable, by “blasting” against GenBank (http://www.ncbi.nlm.nih.gov/) deposited sequences.
Results
Out of a total of 136 species (a total of 164 accessions) on the FLEPPC list across the three gene regions we had a total success of amplification and sequencing at around 99% (see table 1) of complete sampling (Nexus files of the full data sets are freely available on our FTP site and anyone can download these Nexus files at our FTP site (ftp://flmnh.ufl.edu/public/FLEPPC/fleppcmatrices/). The matrices as self-extracting files are also available on the web site devoted to this project (https://www.floridamuseum.ufl.edu/herbarium/research/barcoding/). For the gene region rbcL (ffwcc-uf8162-rbcL.sea.bin), we were able to obtain complete data across all species sampled. For matK (ffwcc-uf8162-matK.sea.bin), data could not be obtained for three samples, all ferns. For the trnH-psbA spacer (ffwcc-uf8162-trnH-psbA.sea.bin), data could not be obtained for five samples: one accession of Colocasia esculenta (but we did obtain the sequence from a second accession of this species), Melia azedarach, Myriophyllum spicatum, Tribulus cistoides, and Xanthosoma sagittifolium. All of these are resolved by the combined rbcL/matK sequences, so this is not a concern to us.
The “problems”
In the collection phase of this project, some species were especially difficult to locate in the wild. For example, Ipomoea aquatica and I. fistulosa could only be procured from horticultural material. Despite being tracked invasives, these species were not readily encountered in the landscape. During the lab process, several problems arose especially during PCR of the DNA region matK. This region proved to be very difficult to amplify using the barcoding primer recommendations made by Kew (http://www.kew.org/barcoding/update.html), with an amplification success around 55%. However, after a literature search for more universal primers, the same portion of matK was amplified more reliably (i.e., ~90%) using primers from Cuenoud et al. (2002). This region also proved difficult to amplify because of the large phylogenetic breadth among the sampled taxa in this study. That is, when one tries to amplify a region that is as variable as matK across all of vascular plants, it is very challenging to get “universal” primers to work consistently. Ferns of the genus Nephrolepis proved especially difficult to sequence; more research will be needed to find matK primers that amplify across all vascular plants (or even all ferns). Preliminarily, we therefore recommend using a combination of primers to have increased success in amplification of matK.
The three accessions of Alstonia macrophylla did not form a clade as sister taxa (with the result that Alstonia does not appear monophyletic), and it seems one of the Alstonia macrophylla accessions (JRA25075), is an unknown entity (morphologically similar to, yet genetically distinct from, A. macrophylla) that is ‘hiding’ in the Miami landscape or that lateral gene flow (e.g., hybridization) with another cultivated Alstonia species (or potentially even a taxon from another genus) has confused the issue. When one considers that there are over 130 species named in Alstonia, it seems this is a likely explanation.
Conclusions/Recommendations
The 100% success rate of obtaining data for all species in this study and the nearly 99% success at obtaining data on a per-region basis provides compelling evidence that barcoding is logistically feasible. Many other published studies on the feasibility of barcoding have focused on which DNA regions to use or in which taxonomic situations might barcoding fail or succeed. Because this data set represents a very broad phylogenetic sampling, we think that this is good evidence that these data can be obtained, analyzed, and compared for a larger sampling of species for the state.
Although not directly informative to the process of barcoding, these data were phylogenetically analyzed as a secondary process for DNA sequence verification. An encouraging result from this analysis is that nearly all of the species are monophyletic where accessions were duplicated. The monophyly of these species sampled further demonstrates the feasibility of barcoding (i.e., synapomorphic DNA characters connote unique sequence variation critical to barcoding efforts). However, because these data will eventually be deposited in an online repository (Consortium for the Barcode of Life, CBOL), this phylogenetic analysis is an unfair test of how the data will be practically used and implemented. CBOL uses a modified “BLAST” analysis to identify an unknown sequence.
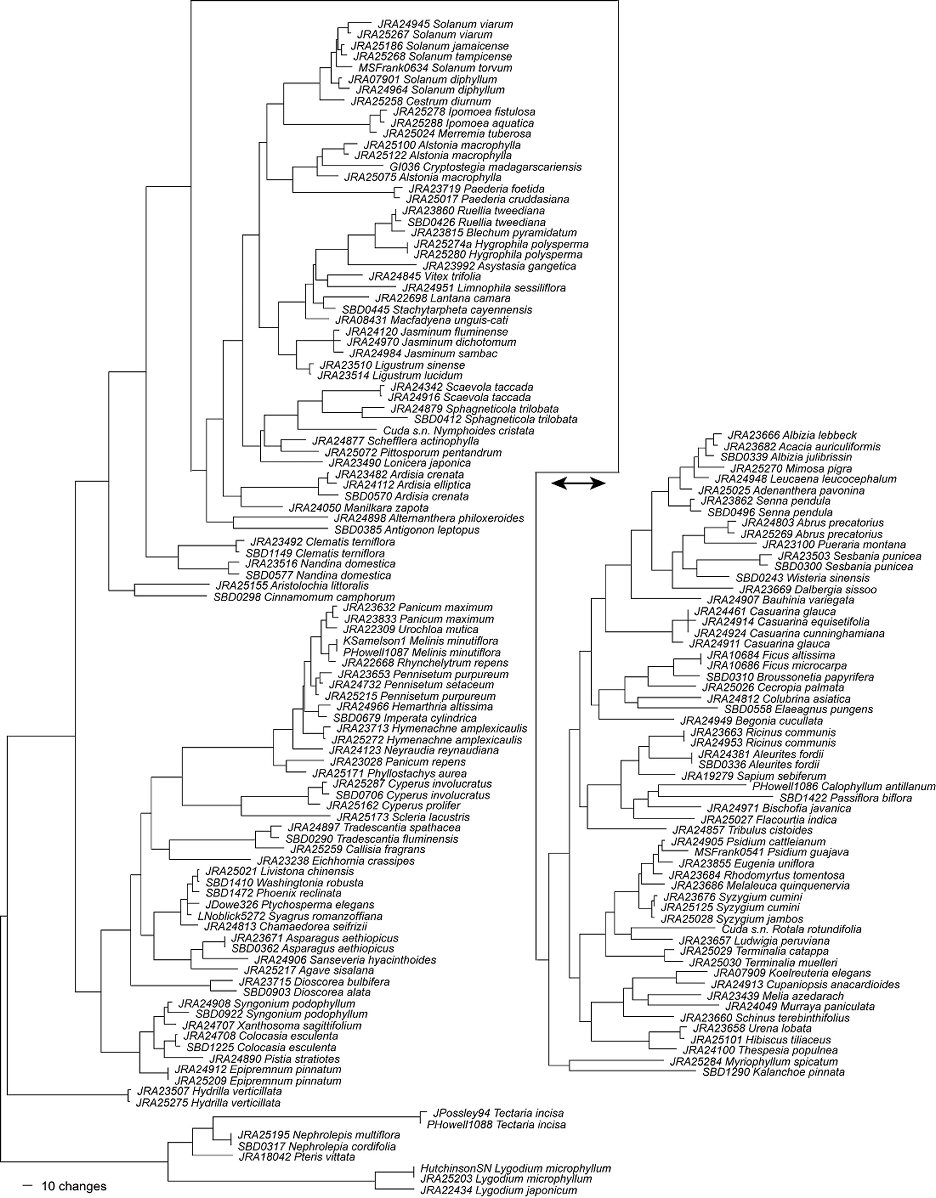
If the FLEPPC species are compared to each other (Figure 1), all of these species can be easily distinguished. Thus any worker with the proper equipment can now sequence a plant suspected of being a FLEPPC listed exotic, with the caveat that 100% identification is not guaranteed. Unfortunately, we do not yet know if these data alone are sufficient to differentiate the invasive plant species from the rest of the flora. If a person were to sequence the barcode DNA regions for an unknown sample and compared the sequence to our dataset of invasive species, the result could be taken as a false positive. Without a complete sampling of all species that occur within Florida, the identification of an unknown sample would be open to interpretation. For example, the two species of Cyperus (C. involucratus and C. prolifer) have many closely related species native to Florida. Because closely related species will inherently have similar DNA sequences, this makes identification of an unknown sequence very difficult without a complete reference of DNA sequences. Without further sampling of Florida plant species, there may be some cases of false positives in identifying FLEPPC taxa, confusing them with closely related taxa. Because of this, we recommend that all plant species in Florida be sequenced for DNA barcoding. The standardized protocols for plant barcoding are still under active discussion and development, and research may eventually produce better barcode regions. With the DNAs of these vouchered specimens in UF archival cryogenic storage, it will be easy to expand upon these studies as plant barcode protocols become more refined.
Future dissemination
In addition to providing sequence data for the previously mentioned three gene regions and vouchers for all material collected, we plan on enhancing these data with additional information. First, we plan to host images of the invasive species (herbarium specimens, and live plant photographs) on the FLAS herbarium website. Second, the data will be submitted to GenBank, the international repository for sequence data. Because the Consortium for the Barcode of Life (CBOL) is not accepting barcoding sequence data for plants at this time, these data have not been submitted to that institution yet. However, when it does go online, we will submit these sequences. And third, all total DNAs will be placed in the cryogenic facility (FLMNH-GRR) in the museum for long term storage, making these available to any researcher in the world, and also providing a ready-made starting point for any future DNA work, including following up on advances in DNA barcoding. Finally, we will continue to build on what we have started with the FLEPPC taxa, working toward DNA barcoding the entire flora of Florida.
Literature Cited
- CHASE, M. W. & H. G. HILLS. 1991. Silica gel: an ideal material for field preservation of leaf samples for DNA studies.Taxon 40: 215-220.
- CUENOUD, P., SAVOLAINEN, V., CHATROU, L. W., POWELL, M., GRAYER, R. J., CHASE, M. W. 2002. Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB, and matK DNA sequences. Am. J. Bot. 89: 132-144.
- DOYLE, J. J., & J. L. DOYLE. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19: 11-15.
- FAZEKAS, A. J., BURGESS, K. S., KESANAKURTI, P. R., GRAHAM, S. W., NEWMASTER, S. G., HUSBAND, B. C., PERCY, D. M., HAJIBABAEI, M., BARRETT, S. C. H. 2008. Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS ONE 3, e2802. http://www.plosone.org/article/info:doi%2F10.1371%2Fjournal.pone.0002802
- FITCH, W. M. 1971. Toward defining course of evolution – Minimum change for a specific tree topology. Systematic Zoology 20: 406-416.
- KRESS, W. J., & D. L. ERICKSON. 2007. A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2(6): e508. doi:10.1371/journal.pone.0000508
- RAMBAUT, A. 1996. Se-Al: Sequence Alignment Editor. Available at http://evolve.zoo.ox.ac.uk/.
- SWOFFORD, D. L. 2003. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods), version 4.0b10. Sinauer, Sunderland, Massachusetts, USA.
Table 1. Vouchers of species used in this study and success for the three DNA regions
(Y = adequate sequence data, NA = no available data). Vouchers are deposited at the UF Herbarium, Florida Museum of Natural History (FLAS), except for two deposited at Fairchild Tropical Garden (FTG).
| Voucher | Species Categories I and II | matK | rbcL | trnH-psbA |
|---|---|---|---|---|
| JR Abbott 24803 | Abrus precatorius | Y | Y | Y |
| JR Abbott 23682 | Acacia auriculiformis | Y | Y | Y |
| JR Abbott 25025 | Adenanthera pavonina | Y | Y | Y |
| JR Abbott 25217 | Agave sisalana | Y | Y | Y |
| SB Davis 0339 | Albizia julibrissin | Y | Y | Y |
| JR Abbott 23666 | Albizia lebbeck | Y | Y | Y |
| JR Abbott 24381 | Aleurites fordii | Y | Y | Y |
| SB Davis 0336 | Aleurites fordii | Y | Y | Y |
| JR Abbott 25075 | Alstonia macrophylla | Y | Y | Y |
| JR Abbott 25100 | Alstonia macrophylla | Y | Y | Y |
| JR Abbott 25122 | Alstonia macrophylla | Y | Y | Y |
| JR Abbott 24898 | Alternanthera philoxeroides | Y | Y | Y |
| SB Davis 0385 | Antigonon leptopus | Y | Y | Y |
| JR Abbott 23482 | Ardisia crenata | Y | Y | Y |
| SB Davis 0570 | Ardisia crenata | Y | Y | Y |
| JR Abbott 24112 | Ardisia elliptica | Y | Y | Y |
| JR Abbott 25155 | Aristolochia littoralis | Y | Y | Y |
| JR Abbott 23671 | Asparagus aethiopicus | Y | Y | Y |
| SB Davis 0362 | Asparagus aethiopicus | Y | Y | Y |
| JR Abbott 23992 | Asystasia gangetica | Y | Y | Y |
| JR Abbott 24907 | Bauhinia variegata | Y | Y | Y |
| JR Abbott 24949 | Begonia cucullata | Y | Y | Y |
| JR Abbott 24971 | Bischofia javanica | Y | Y | Y |
| JR Abbott 23815 | Blechum pyramidatum | Y | Y | Y |
| SB Davis 0310 | Broussonetia papyrifera | Y | Y | Y |
| JR Abbott 25259 | Callisia fragrans | Y | Y | Y |
| P Howell 1086 | Calophyllum antillanum | Y | Y | Y |
| JR Abbott 24924 | Casuarina cunninghamiana | Y | Y | Y |
| JR Abbott 24914 | Casuarina equisetifolia | Y | Y | Y |
| JR Abbott 24461 | Casuarina glauca | Y | Y | Y |
| JR Abbott 24911 | Casuarina glauca | Y | Y | Y |
| JR Abbott 25026 | Cecropia palmata | Y | Y | Y |
| JR Abbott 25258 | Cestrum diurnum | Y | Y | Y |
| JR Abbott 24813 | Chamaedorea seifrizii | Y | Y | Y |
| SB Davis 0298 | Cinnamomum camphora | Y | Y | Y |
| JR Abbott 23492 | Clematis terniflora | Y | Y | Y |
| SB Davis 1149 | Clematis terniflora | Y | Y | Y |
| JR Abbott 24708 | Colocasia esculenta | Y | Y | NA |
| SB Davis 1225 | Colocasia esculenta | Y | Y | Y |
| JR Abbott 24812 | Colubrina asiatica | Y | Y | Y |
| G Ionta 36 | Cryptostegia madagascariensis | Y | Y | Y |
| JR Abbott 24913 | Cupaniopsis anacardioides | Y | Y | Y |
| JR Abbott 25287 | Cyperus involucratus | Y | Y | Y |
| SB Davis 0706 | Cyperus involucratus | Y | Y | Y |
| JR Abbott 25162 | Cyperus prolifer | Y | Y | Y |
| JR Abbott 23669 | Dalbergia sissoo | Y | Y | Y |
| SB Davis 0903 | Dioscorea alata | Y | Y | Y |
| JR Abbott 23715 | Dioscorea bulbifera | Y | Y | Y |
| JR Abbott 23238 | Eichhornia crassipes | Y | Y | Y |
| SB Davis 0558 | Elaeagnus pungens | Y | Y | Y |
| JR Abbott 24912 | Epipremnum pinnatum cv. aureum | Y | Y | Y |
| JR Abbott 25209 | Epipremnum pinnatum cv. aureum | Y | Y | Y |
| JR Abbott 23855 | Eugenia uniflora | Y | Y | Y |
| JR Abbott 10684 | Ficus altissima | Y | Y | Y |
| JR Abbott 10686 | Ficus microcarpa | Y | Y | Y |
| JR Abbott 25027 | Flacourtia indica | Y | Y | Y |
| JR Abbott 24966 | Hemarthria altissima | Y | Y | Y |
| JR Abbott 25101 | Hibiscus tiliaceus | Y | Y | Y |
| JR Abbott 23507 | Hydrilla verticillata | Y | Y | Y |
| JR Abbott 25275 | Hydrilla verticillata | Y | Y | Y |
| JR Abbott 25274a | Hygrophila polysperma | Y | Y | Y |
| JR Abbott 25280 | Hygrophila polysperma | Y | Y | Y |
| JR Abbott 23713 | Hymenachne amplexicaulis | Y | Y | Y |
| JR Abbott 25272 | Hymenachne amplexicaulis | Y | Y | Y |
| SB Davis 0679 | Imperata cylindrica | Y | Y | Y |
| JR Abbott 25288 | Ipomoea aquatica | Y | Y | Y |
| JR Abbott 25278 | Ipomoea fistulosa | Y | Y | Y |
| JR Abbott 24970 | Jasminum dichotomum | Y | Y | Y |
| JR Abbott 24120 | Jasminum fluminense | Y | Y | Y |
| JR Abbott 24984 | Jasminum sambac | Y | Y | Y |
| SB Davis 1290 | Kalanchoe pinnata | Y | Y | Y |
| JR Abbott 07909 | Koelreuteria elegans ssp. formosana | Y | Y | Y |
| JR Abbott 22698 | Lantana camara | Y | Y | Y |
| JR Abbott 24948 | Leucaena leucocephala | Y | Y | Y |
| JR Abbott 23514 | Ligustrum lucidum | Y | Y | Y |
| JR Abbott 23510 | Ligustrum sinense | Y | Y | Y |
| JR Abbott 24951 | Limnophila sessiliflora | Y | Y | Y |
| JR Abbott 25021 | Livistona chinensis | Y | Y | Y |
| JR Abbott 23490 | Lonicera japonica | Y | Y | Y |
| JR Abbott 23657 | Ludwigia peruviana | Y | Y | Y |
| JR Abbott 22434 | Lygodium japonicum | Y | Y | Y |
| Hutchinson s.n. | Lygodium microphyllum | Y | Y | Y |
| JR Abbott 25203 | Lygodium microphyllum | Y | Y | Y |
| JR Abbott 08431 | Macfadyena unguis-cati | Y | Y | Y |
| JR Abbott 24050 | Manilkara zapota | Y | Y | Y |
| JR Abbott 23686 | Melaleuca quinquenervia | Y | Y | Y |
| JR Abbott 23439 | Melia azedarach | Y | Y | NA |
| K Samelson 1 | Melinis minutiflora | Y | Y | Y |
| P Howell 1087 | Melinis minutiflora | Y | Y | Y |
| JR Abbott 25024 | Merremia tuberosa | Y | Y | Y |
| JR Abbott 25270 | Mimosa pigra | Y | Y | Y |
| JR Abbott 24049 | Murraya paniculata | Y | Y | Y |
| JR Abbott 25284 | Myriophyllum spicatum | Y | Y | NA |
| JR Abbott 23516 | Nandina domestica | Y | Y | Y |
| SB Davis 0577 | Nandina domestica | Y | Y | Y |
| SB Davis 0317 | Nephrolepis cordifolia | NA | Y | Y |
| JR Abbott 25195 | Nephrolepis multiflora | NA | Y | Y |
| JR Abbott 24123 | Neyraudia reynaudiana | Y | Y | Y |
| Cuda s.n. | Nymphoides cristata | Y | Y | Y |
| JR Abbott 25017 | Paederia cruddasiana | Y | Y | Y |
| JR Abbott 23719 | Paederia foetida | Y | Y | Y |
| JR Abbott 23632 | Panicum maximum | Y | Y | Y |
| JR Abbott 23833 | Panicum maximum | Y | Y | Y |
| JR Abbott 23028 | Panicum repens | Y | Y | Y |
| SB Davis 1422 | Passiflora biflora | Y | Y | Y |
| JR Abbott 23653 | Pennisetum purpureum | Y | Y | Y |
| JR Abbott 25215 | Pennisetum purpureum | Y | Y | Y |
| JR Abbott 24732 | Pennisetum setaceum | Y | Y | Y |
| SB Davis 1472 | Phoenix reclinata | Y | Y | Y |
| JR Abbott 25171 | Phyllostachys aurea | Y | Y | Y |
| JR Abbott 24890 | Pistia stratiotes | Y | Y | Y |
| JR Abbott 25072 | Pittosporum pentandrum | Y | Y | Y |
| JR Abbott 24905 | Psidium cattleianum | Y | Y | Y |
| M.S. Frank 541 | Psidium guajava | Y | Y | Y |
| JR Abbott 18042 | Pteris vittata | NA | Y | Y |
| John Dowe 326 (FTG) | Ptychosperma elegans | Y | Y | Y |
| JR Abbott 23100 | Pueraria montana var. lobata | Y | Y | Y |
| JR Abbott 23684 | Rhodomyrtus tomentosa | Y | Y | Y |
| JR Abbott 22668 | Rhynchelytrum repens | Y | Y | Y |
| JR Abbott 23663 | Ricinus communis | Y | Y | Y |
| JR Abbott 24953 | Ricinus communis | Y | Y | Y |
| Cuda s.n. | Rotala rotundifolia | Y | Y | Y |
| JR Abbott 23860 | Ruellia tweediana | Y | Y | Y |
| SB Davis 0426 | Ruellia tweediana | Y | Y | Y |
| JR Abbott 24906 | Sansevieria hyacinthoides | Y | Y | Y |
| JR Abbott 19279 | Sapium sebiferum | Y | Y | Y |
| JR Abbott 24342 | Scaevola taccada | Y | Y | Y |
| JR Abbott 24916 | Scaevola taccada | Y | Y | Y |
| JR Abbott 24877 | Schefflera actinophylla | Y | Y | Y |
| JR Abbott 23660 | Schinus terebinthifolius | Y | Y | Y |
| JR Abbott 25173 | Scleria lacustris | Y | Y | Y |
| SB Davis 0496 | Senna pendula var. glabrata | Y | Y | Y |
| JR Abbott 23503 | Sesbania punicea | Y | Y | Y |
| SB Davis 0300 | Sesbania punicea | Y | Y | Y |
| JR Abbott 07901 | Solanum diphyllum | Y | Y | Y |
| JR Abbott 24964 | Solanum diphyllum | Y | Y | Y |
| JR Abbott 25186 | Solanum jamaicense | Y | Y | Y |
| JR Abbott 25268 | Solanum tampicense | Y | Y | Y |
| M.S. Frank 634 | Solanum torvum | Y | Y | Y |
| JR Abbott 24945 | Solanum viarum | Y | Y | Y |
| JR Abbott 25267 | Solanum viarum | Y | Y | Y |
| JR Abbott 24879 | Sphagneticola trilobata | Y | Y | Y |
| SB Davis 0412 | Sphagneticola trilobata | Y | Y | Y |
| SB Davis 0445 | Stachytarpheta cayennensis | Y | Y | Y |
| Larry Noblick 5272 (FTG) | Syagrus romanzoffiana | Y | Y | Y |
| JR Abbott 24908 | Syngonium podophyllum | Y | Y | Y |
| SB Davis 0922 | Syngonium podophyllum | Y | Y | Y |
| JR Abbott 23676 | Syzygium cumini | Y | Y | Y |
| JR Abbott 25125 | Syzygium cumini | Y | Y | Y |
| JR Abbott 25028 | Syzygium jambos | Y | Y | Y |
| J Possley 94 | Tectaria incisa | Y | Y | Y |
| P Howell 1088 | Tectaria incisa | Y | Y | Y |
| JR Abbott 25029 | Terminalia catappa | Y | Y | Y |
| JR Abbott 25030 | Terminalia muelleri | Y | Y | Y |
| JR Abbott 24100 | Thespesia populnea | Y | Y | Y |
| SB Davis 0290 | Tradescantia fluminensis | Y | Y | Y |
| JR Abbott 24897 | Tradescantia spathacea | Y | Y | Y |
| JR Abbott 24857 | Tribulus cistoides | Y | Y | NA |
| JR Abbott 23658 | Urena lobata | Y | Y | Y |
| JR Abbott 22309 | Urochloa mutica | Y | Y | Y |
| JR Abbott 24845 | Vitex trifolia | Y | Y | Y |
| SB Davis 1410 | Washingtonia robusta | Y | Y | Y |
| SB Davis 0243 | Wisteria sinensis | Y | Y | Y |
| JR Abbott 24707 | Xanthosoma sagittifolium | Y | Y | NA |
Figure 1. A phylogram generated from rbcL and matK data generated during this study
(the trnH-psbA spacer was excluded for this figure). Branch lengths are proportional to sequence variation (i.e., the longer the branches, the more likely the taxon is to be distinguishable).