A new paper published today in the journal Nature by an international team of 279 scientists led by the Royal Botanic Gardens, Kew presents the most up-to-date understanding of the flowering plant tree of life.
Using 1.8 billion letters of genetic code from over 9,500 species covering almost 8,000 known flowering plant genera (ca. 60%), this incredible achievement sheds new light on the evolutionary history of flowering plants and their rise to ecological dominance on Earth. The study’s authors believe the data will aid future attempts to identify new species, refine plant classification, uncover new medicinal compounds and conserve plants in the face of climate change and biodiversity loss.
The major milestone for plant science, led by Kew and involving 138 organizations from 27 countries, was built on 15 times more data than any comparable studies of the flowering plant tree of life. Among the species sequenced for this study, more than 800 have never had their DNA sequenced before.
“This is an incredible example of collaboration among the world’s botanists, and the result is new insight into plant evolutionary history,” said co-author Pam Soltis, a distinguished professor and curator at the Florida Museum of Natural History.
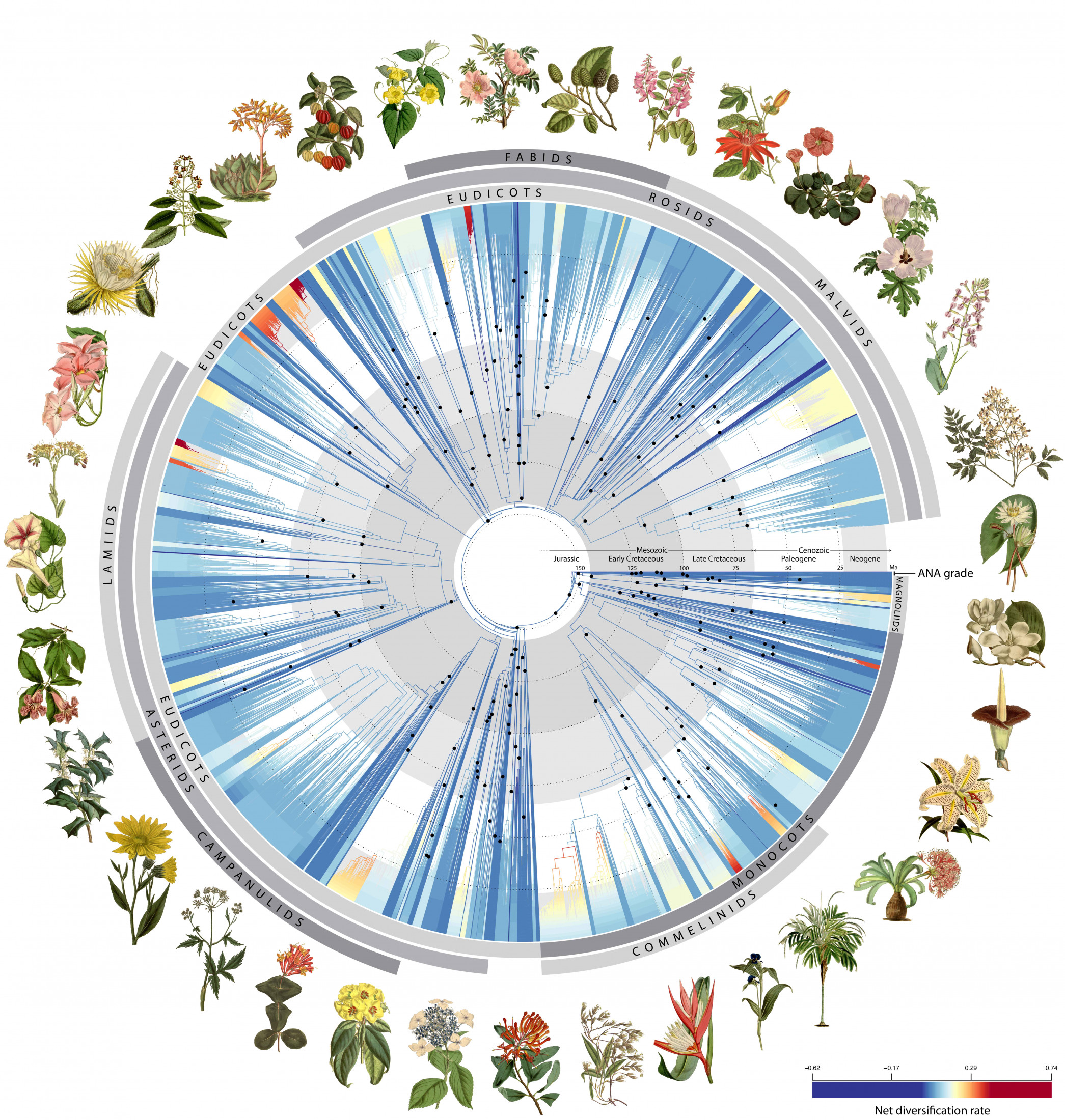
Image by Zuntini et al., 2024, CC BY
Whereas previous studies have often heavily relied on DNA from chloroplasts, the source of energy production in plants, researchers in this study sequenced DNA from nuclei. The latter can be difficult to analyze, but it contains unique information about an organism’s evolutionary history.
“Most of our understanding of flowering plant relationships has come from sequencing portions of the chloroplast genome—this study in contrast provides data from numerous nuclear genes, the Rosetta stone we have long needed for interpreting the evolutionary history of this major part of the tree of life,” said co-author and museum distinguished professor Doug Soltis.
Unlocking historic herbarium specimens for cutting-edge research
Flowering plants alone account for about 90% of all known plant life on land and are found virtually everywhere on the planet – from the steamiest tropics to the rocky outcrops of the Antarctic Peninsula.

Image courtesy of Kew, CC BY
The flowering plant tree of life, much like our own family tree, enables us to understand how different species are related to each other. The tree of life is uncovered by comparing DNA sequences between different species to identify changes (mutations) that accumulate over time like a molecular fossil record. Our understanding of the tree of life is improving rapidly in tandem with advances in DNA sequencing technology. For this study, new genomic techniques were developed to magnetically capture hundreds of genes and hundreds of thousands of letters of genetic code from every sample, orders of magnitude more than earlier methods.
A key advantage of the team’s approach is that it enables a wide diversity of plant material, old and new, to be sequenced, even when the DNA is badly damaged. The vast treasure troves of dried plant material in the world’s herbarium collections, which comprise nearly 400 million scientific specimens of plants, can now be studied genetically. Using such specimens, the team successfully sequenced a sandwort (Arenaria globiflora) collected nearly 200 years ago in Nepal and, despite the poor quality of its DNA, were able to place it in the tree of life.

Image courtesy of Kew, CC BY
More than 500 of the species sequenced for the study are currently at risk of extinction, according to the IUCN Red List. The team also analyzed specimens of species that are already extinct, such as the Guadalupe Island olive (Hesperelaea palmeri), which has not been seen alive since 1875.
“In many ways this novel approach has allowed us to collaborate with the botanists of the past by tapping into the wealth of data locked up in historic herbarium specimens, some of which were collected as far back as the early 19th Century,” said co-author William Baker, a senior research leader at Kew. “Our illustrious predecessors such as Charles Darwin or Joseph Hooker could not have anticipated how important these specimens would be in genomic research today.”
Across all 9,506 species sequenced, over 3,400 came from material sourced from 163 herbaria in 48 countries. Additional material from plant collections around the world, including DNA banks, seeds, living collections, have been vital for filling key knowledge gaps to shed new light on the history of flowering plant evolution. The team also benefited from publicly available data for over 1,900 species, highlighting value of the open science approach to future genomic research.
Putting the plant tree of life to good use
The flowering plant tree of life has enormous potential in biodiversity research. This is because, just as one can predict the properties of an element based on its position in the periodic table, the location of a species in the tree of life allows us to predict its properties. The new data will thus be invaluable for enhancing many areas of science and beyond.
To enable this, the tree and all of the data that underpin it have been made openly and freely accessible to both the public and scientific community, including through the Kew Tree of Life Explorer. The study’s authors believe such open access is key to democratizing access to scientific data across the globe.
Open access will also help scientists to make the best use of the data, such as combining it with artificial intelligence to predict which plant species may have molecules with medicinal potential. Similarly, the tree of life can be used to better understand and predict how pests and diseases are going to affect the plants of a given region in the future. Ultimately, the authors note, the applications of these data will be driven by the ingenuity of the scientists accessing the information.
“Plant chemicals have inspired many pharmaceutical drugs but still have great untapped potential to aid future drug discovery,” said co-author and Kew senior research leader Melanie-Jayne Howes. “The challenge is knowing which to investigate scientifically in the search for new medicines out of the 330,000 flowering plant species.”
Sources: Doug Soltis, dsoltis@ufl.edu;
Pam Soltis, psoltis@flmnh.ufl.edu
Media contacts: Jerald Pinson, jpinson@flmnh.ufl.edu;
Sebastian Kettley, s.kettley@kew.org