With toothlike spines and barbs, thorny skates are just as sharp as their name suggests. But these shark relatives are far more mysterious than they first appear.
Scientists at the Florida Museum of Natural History are using new genetic sequencing technology to understand why current management efforts aren’t reversing the species’ dramatic decline.
Supervised by Gavin Naylor, director of the Florida Program for Shark Research, and funded by the Lenfest Ocean Foundation of the Pew Charitable Trust, postdoctoral researchers John Denton and Shannon Corrigan will analyze the thorny skate genome, targeting its most informative regions and using data from more than 500 skate specimens. Their goal is to determine if multiple thorny skate populations need tailored management strategies and better understand how population sizes may have changed over time.

Illustration by Lindsay Gutteridge
The thorny skate’s ‘conservation conundrum’
Fished for bait and human consumption, thorny skates are flattened fish closely related to sharks and rays. Unlike rays, however, skates lack stinging barbs and spines and pose no danger to humans. But humans are a serious threat to thorny skates.
In 2003, the New England Fisheries Management Council put the Skate Fishery Management Plan into place to combat the species’ decline, Denton said. Though the plan has aided other species of skates, thorny skate numbers continue to drop. Between 2003 and 2016, the number of thorny skates caught by fisheries in U.S. waters decreased by 96%.
“The thorny skate is ‘roommates’ in New England waters with six other skate species,” Denton said. “With the skate management plan, some skates recovered a little bit and others didn’t appear to experience any effects at all from the skate fishing restriction, but the thorny skate still hasn’t recovered. It’s a conservation conundrum.”
Florida Museum image by John Denton
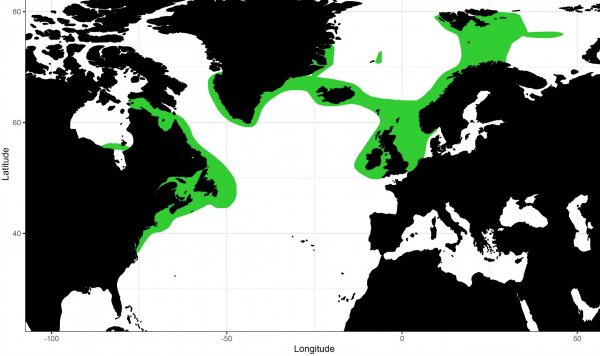
In 2011, organizations began petitioning to list thorny skates as an endangered species based on continued population declines in the Western Atlantic, Denton said. But their conservation status has remained unchanged because previous genetic data suggests thorny skates across the Atlantic are a single population whose numbers can be replenished from the eastern edge of their range.
Researchers noticed that thorny skates appear to mature at a variety of sizes even within the same areas, something that’s not showing up in their DNA and could signal there are multiple distinct populations. But Corrigan said that while tagging data shows thorny skates are relatively sedentary, East and West Atlantic skate populations thousands of miles apart are genetically similar.
“When you look at the genetic data available for the species, you can’t tell skates that vary in size apart,” she said. “Additionally, from existing DNA data it seems like thorny skates at all locations are genetically similar, which is in stark contrast with what we would expect based on the fact that when we tag them they don’t seem to move very far.”
Technology can provide a clearer picture of skate DNA
Now, the Florida Museum shark research program is using the latest sequencing technology to take a higher-resolution look at thorny skate DNA. By determining whether or not there are multiple, non-breeding populations of thorny skates in the Atlantic, scientists will be able to more confidently decide if these populations require individual conservation strategies.
Previous research relied on DNA barcoding, a process in which a region of DNA considered most likely to differ between species is selected for sequencing. But DNA barcoding isn’t always effective. Scientists can’t be certain this technique will yield the right – or even enough – information to distinguish slight differences between populations within a species.
Today, scientists are able to ask questions first and then “fish” for the answers in DNA with specially designed probes, Corrigan said, using a technique called DNA hybridization target sequence capture.
“At the beginning of the project, we’re thinking about the questions that we want to be able to answer and then targeting the regions of the genome that we think will answer those questions,” she said.
“Now, we can develop sophisticated protocols tailored to the questions we’re asking before we begin sequencing,” Denton said. “Before, we relied on generic DNA regions and would hope they worked for the questions we wanted to address.”
Using a complete sequence of thorny skate DNA from the Vertebrate Genomes Project, an initiative that aims to sequence the full genomes of 70,000 vertebrate species, Corrigan and Denton will design probes, similar to how Velcro works, to latch onto specific areas of the genome researchers are interested in studying. The probes are pulled out with a magnetic molecule, with the desired DNA region attached, providing an isolated piece of informative DNA.
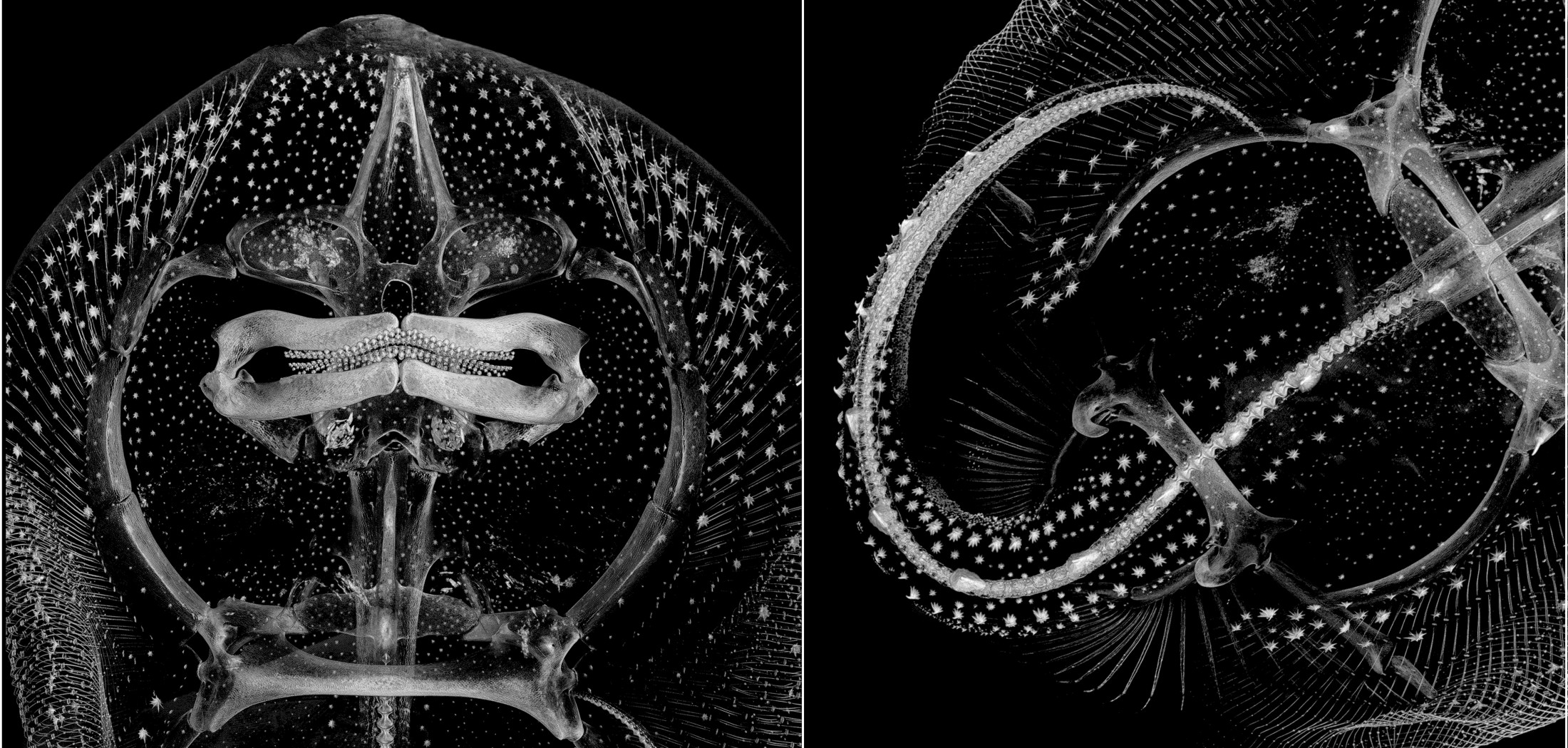
Florida Museum image by Zachary Randall
“Why are we putting so much emphasis on getting good quality molecular information?” Denton said. “The thorny skate problem isn’t hypothetical, it’s not in the future and it’s not retrospective – it’s a current thing. We’re leveraging this genetic information and these techniques in ways that we couldn’t if we were just pulling these little fragments from barcodes.
“I think it’s starting to come to the forefront that more data is good, but more of the right kind of data is best,” he added. “Having high quality genetic data is something that really means you can continue to answer a variety of questions later on.”
Using the target sequence capture method, Denton and Corrigan will focus on the parts of the genome with the most information. When viewing these selections in finer detail, they can discover variations in the genetic sequences that might not have been visible using the barcoding method and determine if there are independent populations of thorny skates.
“The thorny skate conundrum is a known issue,” Denton said. “Previous genetic information was the major justification for not listing thorny skates with any distinct population segments that would need different conservation recommendations.”
“We’ve had all the information we could have up until now, but we still weren’t sure what was really going on, so it’s been hard to make decisions we were confident in simply because we only had access to a limited amount of information,” Corrigan said. “Now, we have these new tools that allow us to ask informed questions at the outset and that will allow us to manage species that are being pressured in a more effective way.”
In addition to explaining the size differences in thorny skates, new DNA data could also reveal whether there’s a “cryptic species” involved – an animal that physically appears identical to the thorny skate but has different DNA.
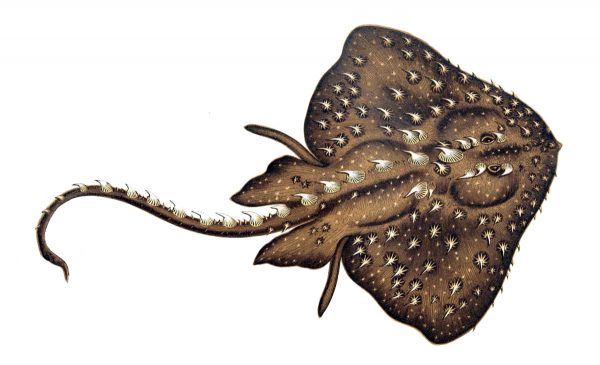
Illustration by Henri Gervais in the public domain
“One of the immediate things with genetic information is that it can show variation that’s otherwise invisible,” Denton said. “If you’re just looking at the organism, they may all look similar in any way you could visually find to diagnose them, but there could be significant genetic differences.”
The shark research team is using thorny skate samples from across the Atlantic, obtained with help from collaborators at the New England Aquarium, the Marine and Freshwater Research Institute in Iceland, the Norwegian Institute of Marine Research and the Arctic University of Norway.
“It’s a nice kind of synergy of all these different things coming together,” Corrigan said. “We now can have really good sampling going on throughout the range of thorny skates thanks to our collaborators and the fisheries work that’s already been done in addition to the advances in the field of genomics over the last couple of decades.”
The team plans to share its data and recommendations with interested fisheries, including eventually uploading it to GenBank, a public repository for genetic data.
“We’re hoping the thorny skate pipeline will be used as a template for using sequencing technologies in the future for fisheries conservation recommendations and communication between fisheries folks and population biologists,” Denton said.
“It’s going to have a real-world outcome and a practical management impact,” Corrigan said. “Beyond this project, this will be a genomic resource available for thorny skates forever.”
- Learn more about the project.
- Learn more about the Chondrichthyan Tree of Life.
- Learn more about thorny skates.
Sources: John Denton, jdenton@floridamuseum.ufl.edu;
Shannon Corrigan, scorrigan@floridamuseum.ufl.edu