Marchant, D. B., E. B. Sessa, P. G. Wolf, K. Heo, W. B. Barbazuk, P. S. Soltis, and D. E. Soltis. 2019. The C-Fern (Ceratopteris richardii) genome: insights into plant genome evolution with the first partial homosporous fern genome assembly. Scientific Reports 9. [View on publisher’s site]
Abstract
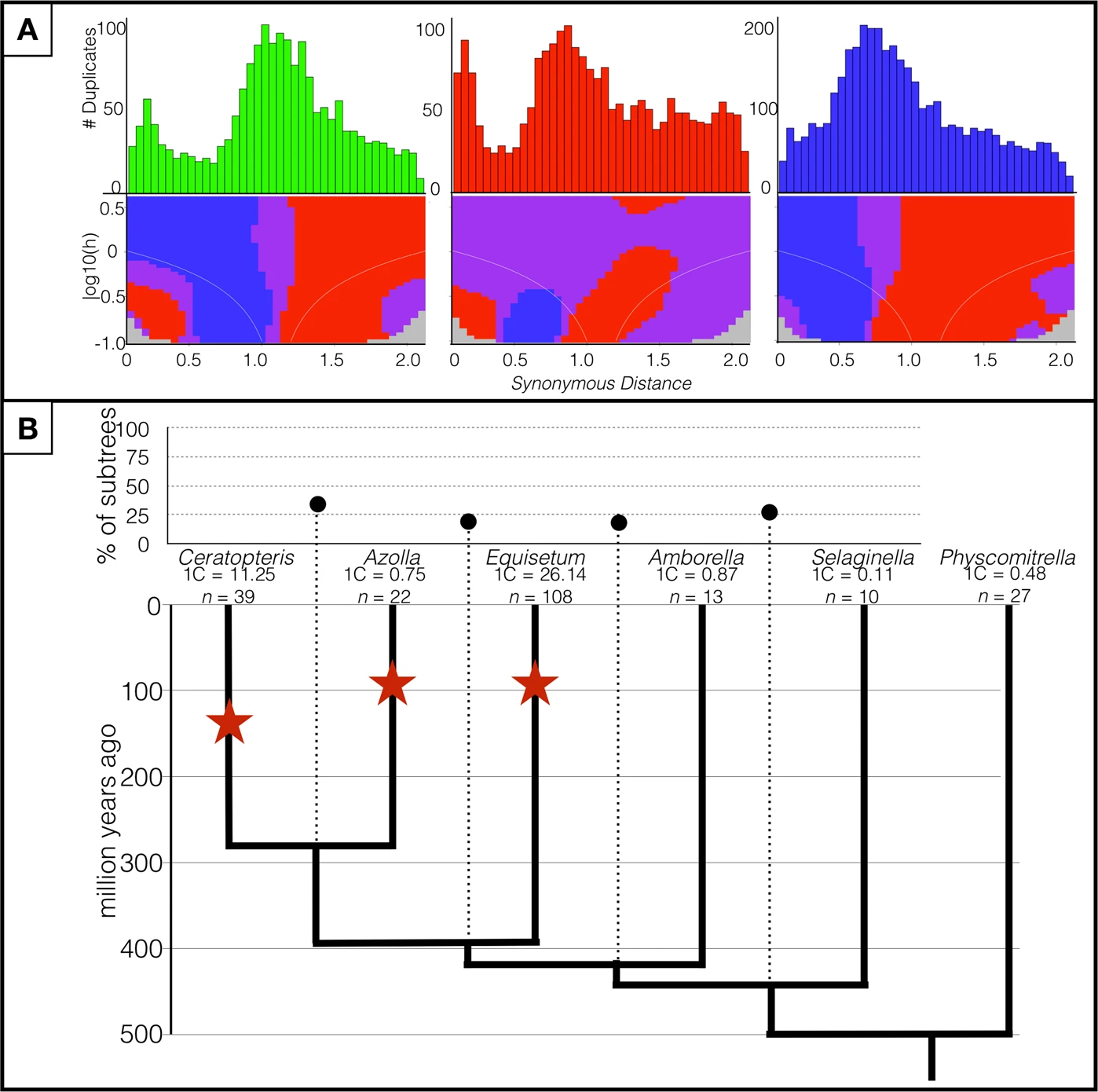
Ferns are notorious for possessing large genomes and numerous chromosomes. Despite decades of speculation, the processes underlying the expansive genomes of ferns are unclear, largely due to the absence of a sequenced homosporous fern genome. The lack of this crucial resource has not only hindered investigations of evolutionary processes responsible for the unusual genome characteristics of homosporous ferns, but also impeded synthesis of genome evolution across land plants. Here, we used the model fern species Ceratopteris richardii to address the processes (e.g., polyploidy, spread of repeat elements) by which the large genomes and high chromosome numbers typical of homosporous ferns may have evolved and have been maintained. We directly compared repeat compositions in species spanning the green plant tree of life and a diversity of genome sizes, as well as both short- and long-read-based assemblies of Ceratopteris. We found evidence consistent with a single ancient polyploidy event in the evolutionary history of Ceratopteris based on both genomic and cytogenetic data, and on repeat proportions similar to those found in large flowering plant genomes. This study provides a major stepping-stone in the understanding of land plant evolutionary genomics by providing the first homosporous fern reference genome, as well as insights into the processes underlying the formation of these massive genomes.